DNA Technology

Re-inventing DNA
As we search for life in the cosmos, we must ask which molecular details of the Terran biosphere are general to life universally, and which reflect historical accidents and contingencies. Synthesis offers a way to answer such questions. Using organic chemistry tools, our group synthesized many molecular systems that are not DNA or RNA, but can store information like DNA and RNA. More importantly, we synthesized many molecular systems that cannot store information like DNA/RNA. Study of these alternative candidates for genetic material provides the experimental grounds for the agnostic life finder (ALF) that is being prepared for flight. With researchers at Indiana University, crystal structures of many of these alternative life forms are being solved. The National Science Foundation is presently supporting this work.
Examples of re-invented DNA and RNA include:
- Artificially expanded genetic information systems (AEGIS), which contain as many as 12 different nucleotide building blocks.
- Combinations of these molecules, including "fat" and "skinny" DNA.
- Self-avoiding molecular recognition systems (SAMRS), which are especially valuable for multiplexed diagnostics.
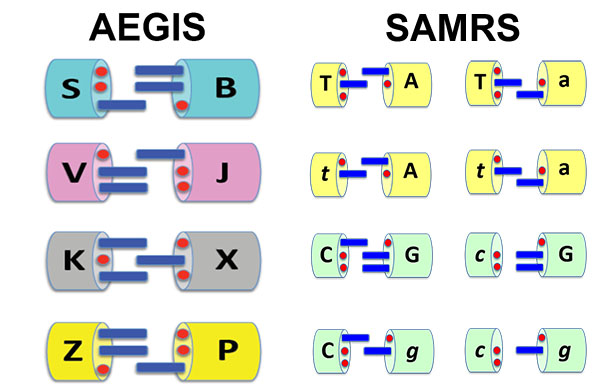
The hydrogen bonding patterns of artificially expanded genetic information systems (AEGIS) and self-avoiding molecular recognition systems (SAMRS).



